Hi @vickylouise,
I was thinking I could calibrate each channel separately by connecting the photodiode to the REF and 90 pockets to calibrate 90, and the REF and 45 to calibrate 45 but then we’d have two calibrations and I believe only one can be active at a time. Do you have any suggestions?
Unfortunately this won’t work, as the calibration will be scaled by the REF, but in practice, it won’t be, so you won’t be “mapping” to the correct spot.
As a simple first step to fix the above, I suggest copy-pasting the protocol you want (singe-vial vs standards, I use single-vial below), making the following edits, and adding it as a plugin in ~/.pioreactor/plugins.
Changes to make:
-
Change:
if "REF" not in config["od_config.photodiode_channel_reverse"]:
echo(
red(
"REF required for OD calibration. Set an input to REF in [od_config.photodiode_channel] in your config."
)
)
raise click.Abort()
# technically it's not required? we just need a specific PD channel to calibrate from.
ref_channel = config["od_config.photodiode_channel_reverse"]["REF"]
pd_channel = cast(pt.PdChannel, "1" if ref_channel == "2" else "2")
to
pd_channel = prompt(
green("Provide the PD channel you are calibrating"),
type=click.Choice(["1", "2"])
)
-
At the bottom of that file, add the following:
from pioreactor.calibrations import CalibrationProtocol
class MultiplePDCalibration(CalibrationProtocol):
target_device = "od"
protocol_name = "single_vial_multiple_pd"
def run(self):
return run_od_calibration()
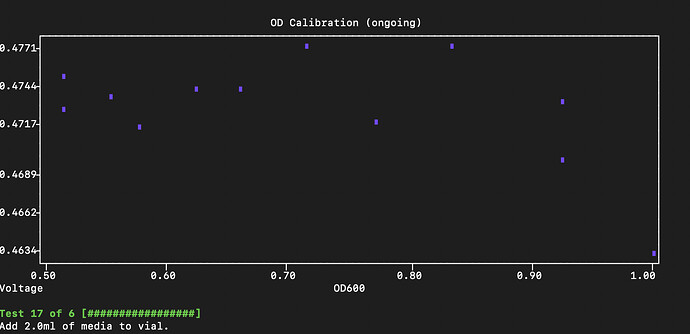
Now, I think, when you run pio calibrations run --device od --protocol-name single_vial_multiple_pd, your code will execute and and app will let you specify the PD you want to use for the calibration.
That allows you to create calibrations per PD. If everything works, you should see them in the folder ~/.pioreactor/storage/calibrations/od/ with different pd_channel fields.
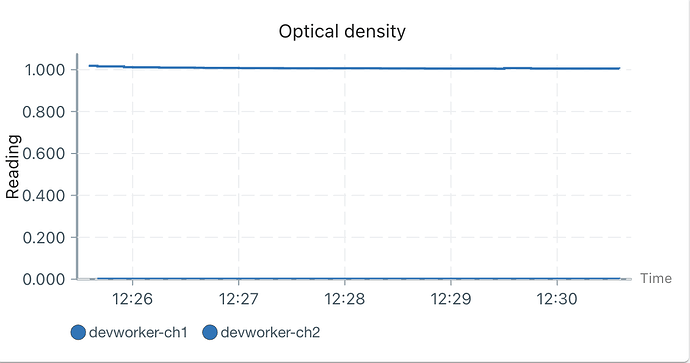
Next, we need to modify od_reading to allow for two calibrations. You’re correct that only a single active calibration is possible, but we can get around this. The calibration logic in od_reading is setup to have a calibration per PD, see self.models in this code.
Anyways, a new plugin Python file the following would work (TODO: fill in the TODOs in the file below):
# -*- coding: utf-8 -*-
from __future__ import annotations
from pioreactor.background_jobs.od_reading import *
from pioreactor.calibrations import load_calibration
def start_od_reading(
od_angle_channel1: pt.PdAngleOrREF
| None = cast(pt.PdAngleOrREF, config.get("od_config.photodiode_channel", "1", fallback=None)),
od_angle_channel2: pt.PdAngleOrREF
| None = cast(pt.PdAngleOrREF, config.get("od_config.photodiode_channel", "2", fallback=None)),
interval: float | None = 1 / config.getfloat("od_reading.config", "samples_per_second", fallback=0.2),
fake_data: bool = False,
unit: str | None = None,
experiment: str | None = None,
):
if interval is not None and interval <= 0:
raise ValueError("interval must be positive.")
if od_angle_channel2 is None and od_angle_channel1 is None:
raise ValueError("Atleast one of od_angle_channel2 or od_angle_channel1 should be populated")
unit = unit or whoami.get_unit_name()
experiment = experiment or whoami.get_assigned_experiment_name(unit)
ir_led_reference_channel = find_ir_led_reference(od_angle_channel1, od_angle_channel2)
channel_angle_map = create_channel_angle_map(od_angle_channel1, od_angle_channel2)
channels = list(channel_angle_map.keys())
# use IR LED reference to normalize?
if ir_led_reference_channel is not None:
ir_led_reference_tracker = PhotodiodeIrLedReferenceTrackerStaticInit(
ir_led_reference_channel,
)
channels.append(ir_led_reference_channel)
else:
ir_led_reference_tracker = NullIrLedReferenceTracker()
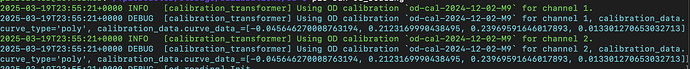
calibration_pd1 = load_calibration("od", ) # TODO: PROVIDE NAMES OF CALIBRATIONS
calibration_pd2 = load_calibration("od", ) # TODO: PROVIDE NAMES OF CALIBRATIONS
calibration_transformer = CachedCalibrationTransformer()
calibration_transformer.hydate_models(calibration_pd1)
calibration_transformer.hydate_models(calibration_pd2)
if interval is not None:
penalizer = config.getfloat("od_reading.config", "smoothing_penalizer", fallback=700.0) / interval
else:
penalizer = 0.0
return ODReader(
channel_angle_map,
interval=interval,
unit=unit,
experiment=experiment,
adc_reader=ADCReader(
channels=channels, fake_data=fake_data, dynamic_gain=not fake_data, penalizer=penalizer
),
ir_led_reference_tracker=ir_led_reference_tracker,
calibration_transformer=calibration_transformer,
)
@click.command(name="od_reading")
@click.option(
"--od-angle-channel1",
default=config.get("od_config.photodiode_channel", "1", fallback=None),
type=click.STRING,
show_default=True,
help="specify the angle(s) between the IR LED(s) and the PD in channel 1, separated by commas. Don't specify if channel is empty.",
)
@click.option(
"--od-angle-channel2",
default=config.get("od_config.photodiode_channel", "2", fallback=None),
type=click.STRING,
show_default=True,
help="specify the angle(s) between the IR LED(s) and the PD in channel 2, separated by commas. Don't specify if channel is empty.",
)
@click.option("--fake-data", is_flag=True, help="produce fake data (for testing)")
@click.option("--snapshot", is_flag=True, help="take one reading and exit")
def click_od_reading(
od_angle_channel1: pt.PdAngleOrREF, od_angle_channel2: pt.PdAngleOrREF, fake_data: bool, snapshot: bool
) -> None:
"""
Start the optical density reading job
"""
if snapshot:
od = start_od_reading(
od_angle_channel1,
od_angle_channel2,
fake_data=fake_data or whoami.is_testing_env(),
interval=None,
)
od.logger.debug(od.record_from_adc())
# end early
return
else:
od = start_od_reading(
od_angle_channel1,
od_angle_channel2,
fake_data=fake_data or whoami.is_testing_env(),
)
od.block_until_disconnected()
This will overwrite the usual pio run od_reading with this new code. Does that make sense?
There’s another solution that would involve creating new “calibration devices”, one per PD. So, instead of od as the device, it’s od_pd1 and od_pd2. Then you can have again a single active calibration per device. This is a pretty good solution, too. It would require more custom calibration code though.