It would be nice to have some way of re-normalizing our OD through the UI. Maybe we could have an additional page called “Media” and it could let us save and load different Media recipes, perform calibrations, and display data (e.g., machine specific calibration data, or user entered data).
I saw you’re planning something like this. Do you have an idea of when this might be available, and can you share any details about what this might look like?
Glad you’re interested in it! The CLI app that currently exists is a temporary solution. CLI apps are pretty quick to build, but aren’t very accessible or scalable. We do have plans to move control into the UI, along with other calibrations (pump, stirring, etc.)
There’s a bit of technical backend work that needs to be done first¹. That’s not too hard, but takes some time. The real difficulty is the UI: it’ll be quite a complex interface to design and code up.
So it’s a feature on our roadmap, but will take on the order of months, easily. Once the backend is done, we can start to introduce small adjustments to the UI (ex: launching with the ability to display existing calibrations and choose one). Later, we’ll introduce running calibrations from the UI, editing calibrations, etc.
¹ Specifically, the difficulty is keeping things in sync: if you change a calibration on a worker, the leader (UI) needs to be updated, and vice versa. So we need to consider a protocol/message passing between the leader (UI) and the worker.
Thanks, good to know.
Is there a way I could try and calibrate it whenever I start an experiment? So far I haven’t been doing much to control what my nOD reference point is.
Here’s what I’m thinking. When I set up an experiment, I can prepare the media as I usually do, but I can also pour some into an extra 20 mL vial and cap it immediately after sterilization. When I start my new experiment, I can insert this vial first, start up the OD Reader and Growth Rate jobs, let them determine an initial reference point, and then when the initial calculations are finished, I can pause both jobs, replace the sample vial with the culture chamber, inoculate the experiment, and then resume the OD Reader and Growth Rate jobs.
Also, I’m looking at the page on OD600 Calibration, but I’m not sure if that is what I’m trying to do. I’m usually making changes to media from experiment to experiment. I want to run some one-off experiments, and this calibration seems like it is designed for more repeatable experimental methods. Can you also maybe explain a bit of what is being calibrated as well?
Are you looking to record blanks? Setting up an experiment | Pioreactor Docs
This is for measuring the baseline measurement of the media, prior to any inoculation.
Thanks, yeah that was what I was looking for. Didn’t realize that button was there.
Nevermind, I think I’m asking about something a little different.
I measured the Blank OD for some media I have and it was 0.03~0.05. When I start an experiment, after inoculating the yeast and starting the growth rate job, my reference OD might get set at 0.2 or 0.3. My maximum population usually reaches at least 0.8 to 1.2. Also, if I want to start multiple experiments, I want to have them using the same reference sample. Because my measured OD readings are so much higher, I’m not too worried about the offset from the Blank. I want to set my reference OD to be equal to the Blank OD.
IIUC, in your system, take for example, an nOD of 2.5 represents the culture turbidity is 2.5x the reference’s turbidity (blank media perhaps). If the culture doubles, the nOD rises to 5.0, and that represents a 100% growth.
In the current system, the reference is the starting OD, so we get the nice ratio of nOD = 1.0. If the culture doubles, the nOD rises to 2.0, and that represents a 100% growth.
Thinking through this, I think everything should work, including growth rate calculations, if you choose a different reference. You do lose the following interpretation of the nOD: the nOD represents the culture’s multiplicative growth (ex: nOD of 2.0 represents a doubling, etc.)
The strategy of pausing and replacing the vial is iffy. Things will probably blow up. Let me think about this a bit more, and develop a strategy. Something like a config.ini parameter for growth_rate_calculating that if set to true will use a reference value instead of calculating it inline (as it currently does)
Just making some notes for myself later: growth_rate_calculating looks for a value in the cache od_normalization_mean here, and uses this as the normalization value / reference value. (If it’s not found, we compute the reference as per usual with N samples). A job could place a value here beforehand. Ex: an action could perform something like od_blank, and place the value in the cache (this suggests a plugin). No modification would need to be done to growth_rate_calculating. We may also need to populate the od_normalization_variance, but this could still use the “traditional path” (compute from N samples).
@realPeteDavidson I think this is the plan: a plugin can be created (see below) that will populate the storage that growth_rate_calculating looks at to determine the reference value. By default, if the storage is empty, we compute it as normal. So by running this plugin while just media is in the vial, i.e. before inoculating, it will use the media as a reference.
The plugin starts both stirring and od reading, so these need to be off when the job starts. By default, the plugin will read 30 samples, take an average, and then use this as the reference OD. This can be changed in the code. The job will turn off when finished.
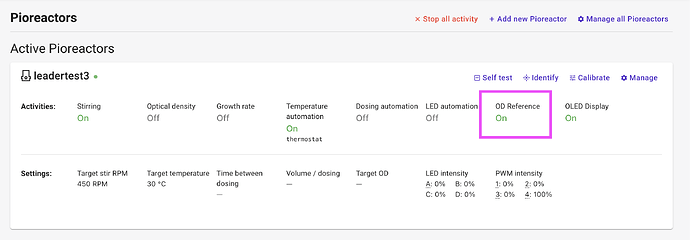
Here’s a link to the Python file and the YAML file to display in the UI: od_reference.py · GitHub
Screen shot of it active:
(Note that Optical density and Stirring will display “Off” in the UI, but they will be running while OD Reference is “On”)
1 Like
Thanks. I’ll let you know how it works when I try it out.
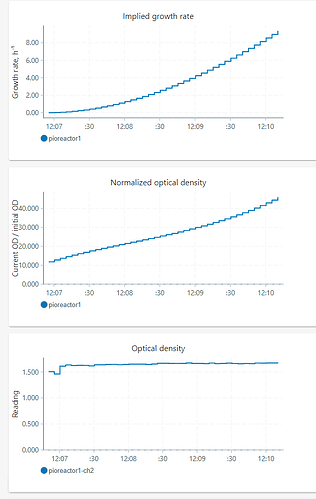
I tried using and the growth/NOD blew up. I started a new experiment, put a reference sample in (media, ~0.003-4 OD), ran the OD Reference script, exchanged the media sample for a previous experiment with ~1.5 OD. When I started the OD_reader, stirrer, and growth rate calculator, the NOD and growth rates blew up. Image below.
hm, okay, I’ll have to test what’s going on internally more. Thanks for the report.
Edit: the problem is this line. The Kalman Filter needs an initial “state” for the nOD, and historically 1.0 has been a good guess for this initial nOD (by construction). However, when we start passing in the observed nOD as in your experiment, it is passed in 1.5 / 0.003 = ~500. So the model thinks “hey I’m at 1.0, but I’m observing 500, I must be growing very fast!”.
Anyways, the solution is remove this hardcoded 1.0 in that line of code.
As I under the Kalman Filter wiki, it looks like the Kalman filter uses the previous internal state plus its measured new internal state to calculate its new state. When starting up GrowthRateCalculator, you need to set some value for the systems initial conditions. Normally Growth Rate Calculator takes a couple minutes to collect some initial data. Could this initial startup period and sample data be used to calculate the initial internal state?
I think this is what you were talking about when removing the hardcoded initial nOD, but I’m curious about whether the initial condition for Growth Rate (which should be part of the internal state I believe) is also hardcoded.
Edit: It might be nice to have the kalman filter ‘reset’ every time you start the growth calculator by collecting some new sample data, or maybe a button that will tell it to restart from initial conditions. I was thinking of letting one colony grow to max, filling another 20mL vial of nutrients, and inoculating it with the original colony.
On a related note, I’ve noticed that running the pumps to add or remove waste seems to influence the growth rate and nOD calculations by causing it to drop, even when they aren’t connected to the chamber. Do these calculations take into account how much media/alt_media/waste the pi thinks it just exchanged?
On a related note, I’ve noticed that running the pumps to add or remove waste seems to influence the growth rate and nOD calculations by causing it to drop, even when they aren’t connected to the chamber. Do these calculations take into account how much media/alt_media/waste the pi thinks it just exchanged?
During a dilution, growth_rate_calculating will adjust some internal parameters to allow for more flexibility. If it didn’t, it’s kinda the same problem as I mentioned above: “i’m at nOD = 2.0, but i’m observing nOD = 1.5 [due to dilution], wow I must be growing really negatively!”. The original paper the algorithm is based on has more specifics. It’s implemented as a MQTT callback listening to dosing_events topics: pioreactor/pioreactor/background_jobs/growth_rate_calculating.py at master · Pioreactor/pioreactor · GitHub
Why is it dropping, though? It’s probably an unintended side-effect, I’ll look into it more.
Normally Growth Rate Calculator takes a couple minutes to collect some initial data. Could this initial startup period and sample data be used to calculate the initial internal state
Yea, I think this is how we’ll proceed. We intentionally skip this collecting in the plugin above, but we’ll add some collecting back in growth_rate_calculating to better initialize nOD.
w.r.t to the initial state of growth_rate: it is hardcoded to 0. This one is harder to predict an initial state for.
I’ve only noticed this behavior occasionally. I’m not usually running pumps or cycling media that are disconnected from a chamber that has yeast growing while the growth rate calculator is running. If the dilutions allow for more flexibility, then the pumps probably just allowed it to adjust quicker. I think the additional flexibility probably explains the behavior I saw. I just remember it caused a sharp discontinuity in the graph, but not whether the graph had already been changing.