Hey!
i’ve recently updated my cluster to 25.5.1 all the way from 24.10.1 (incrementally of course!) to take advantage of the OD600 calibration.
I was using the single vial protocol and measuring OD600 in a spectrophotometer (blanked with Lennox Broth (LB)) whenever prompted.
I’ve tried to run this calibration protocol a few times but it always seems to fail when I get to the end, where I’m asked to provide the OD of my media.
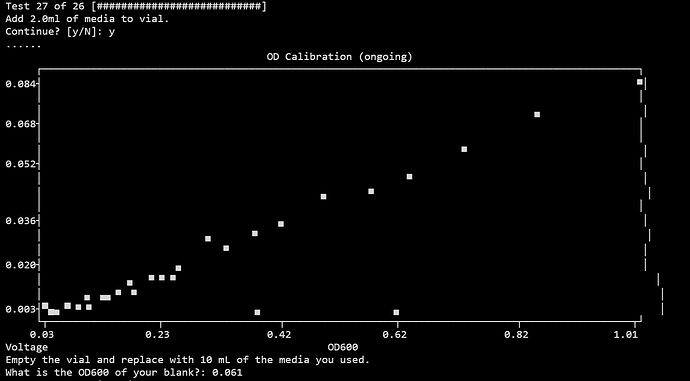
At first I assumed the best way to do this would be to blank my spectrophotometer with water, then measure the OD600 of LB (silly on my part, but I was not too sure what was being asked of me). I entered this value (~0.061), which resulted in the following:
Reading blank...
Reading is too high, trying again...
Reading is too high, trying again...
Reading is too high, trying again...
Reading is too high, trying again...
Reading is too high, trying again...
Why is the blank reading, 0.007061387502738081V, higher than everything else: [0.08381068184289237, 0.07215648153036494, 0.059990495649226594, 0.05049483620097249, 0.04423651213430242, 0.004209839326557134, 0.04119455321826533, 0.03336294465594032, 0.02946311569797438, 0.025674508836002266, 0.003870240992258537, 0.02760270349113583, 0.018774914875512375, 0.01613189389916026, 0.014452835004165283, 0.01590998482902551, 0.013372906049932187, 0.010782692998190914, 0.009790551716845626, 0.008856914993584195, 0.009094404629279238, 0.008290732050476476, 0.0070388222592080085, 0.006004920923917221, 0.005065548401484521, 0.005502664548161679, 0.005096614296223755, 0.0038720353411301565, 0.003920031518861105, 0.003423386589185564, 0.0037728271909961706, 0.004304656479406765, 0.005460199925158296, 0.005269595377488636]V?
Aborted!
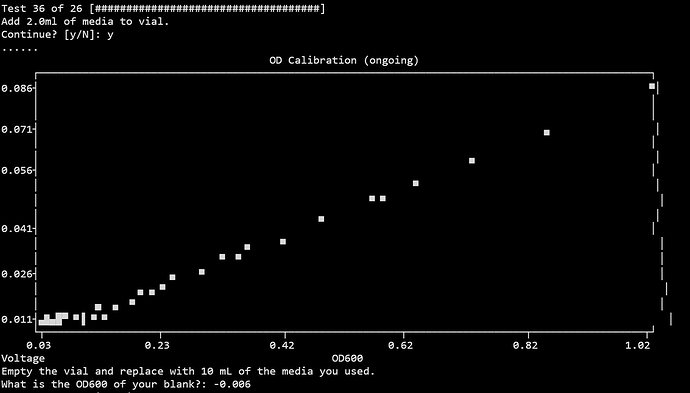
So I repeated the entire process but this time when I was asked to provide the media OD, I kept the spectrophotometer blanked with LB and remeasured a sample of the same LB. = -0.006… basically zero, and definately lower than my other OD readings. Resulted in the same:
Reading is too high, trying again...
Reading is too high, trying again...
Reading is too high, trying again...
Reading is too high, trying again...
Reading is too high, trying again...
Why is the blank reading, 0.011804623729992577V, higher than everything else: [0.08572887852644015, 0.06913144112255011, 0.06055490458238791, 0.05343759899909553, 0.04884866447429399, 0.050517903532944566, 0.04250005741519082, 0.036254001121569984, 0.0331974130194098, 0.032081688791327445, 0.03146681145075177, 0.02784498121982694, 0.024038525857615747, 0.02209818950719277, 0.020272632813524847, 0.020731640539264115, 0.018113063005095027, 0.016773509531510813, 0.01545209817316242, 0.014024421185487917, 0.014980939113016057, 0.014124397378338177, 0.013436110830258531, 0.012556146641814607, 0.012526279286657657, 0.012476324362991481, 0.012708805337619988, 0.012860070246445247, 0.012968241865586785, 0.012190359031105035, 0.01266155225709167, 0.012146777486722512, 0.012030540999525217, 0.011592140435842562, 0.011631321204746416, 0.012906408419762342, 0.011877456060977131, 0.011984718381705617, 0.0115640098727537, 0.011383139760435099, 0.01305428554705593, 0.012505476041388491, 0.013109017645274711, 0.011919741775265492, 0.012259949400437106]V?
Aborted!
I know it could be a case of the first obviously failing because the blank value I provided was very high, and the second was negative. But writing zero here feels equally incorrect so I’m a bit stuck.
A few other bits:
- The curve for attempt 1 looked normal, though there were a few outliers.
- The second attempt was estimated to need 26 2mL dilutions, it actually required 36… So I assumed there may be an issue with the LEDs or something. I ran a self test and everything was normal. I also tried changing the REF value as Cameron outlined here: PD Blank signal too high - #3 by sharknaro, everything worked normally. The curve looked normal but with way more data points.
So, any advice on how to successfully calibrate the OD would be really appreciated!