I didn’t see a forum category for posting and updating experiments, maybe that should be another category separate from this one?
After a basic test run ~2 weeks ago, I decided to try posting my second experiment run here. I am still developing a process for setting up and running an experiment, so I would welcome any suggestions.
Goal: Evolve a colony of yeast in different conditions to see what happens, develop an experimental process for future experiments, and to identify opportunities for improvement.
Organism Strain: Fleischmann’s RapidRise Instant Yeast strain
Nutrient Media: LonoLife Bone Broth 15g + ~10 Oz water (tap)
Alternate Media: ~6.1 molar saltwater (~87g NaCl + ~270mL tap water)
Experimental Method
I plan on running this experiment for ~1-2 weeks. When I refer to an “experiment-day,” I am referring to a 24 hour period starting from whenever the experiment started. If I start an experiment at 8PM Monday, 1 full experiment-day would from 8PM Monday to 8PM Tuesday.
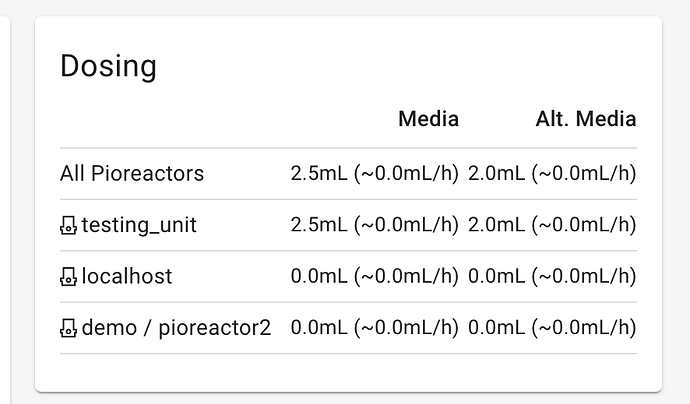
Nutrient Solution: Nutrients are added regularly at a rate of 0.5 ml every 20 minutes, which is 1.5 ml every hour.
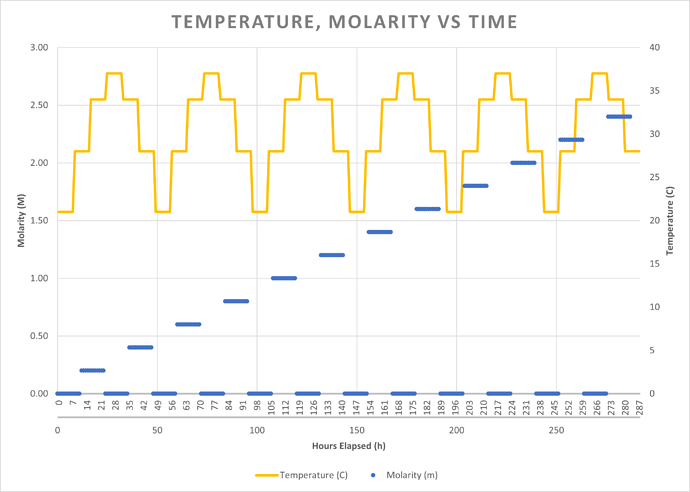
Salt Solution: Saltwater is on a half-on, half-off schedule, where the first 12 hours of any experiment-day have no saltwater added, and the second 12 hours of any experiment-day have some amount of saltwater added regularly. The dose of saltwater added, and the interval between doses, is calculated based upon a desired molarity and the amount of nutrients added every hour (i.e., a constant 1.5ml/hr, for now…). The desired molarity starts at 0.2M and increases by 0.2M every experiment-day.
Example: At hour 13, I want to add saltwater. Because it is experiment-day 1, the desired molarity is 0.2M. To achieve this, the script will add approximately 0.052 ml of 6.0M saltwater every hour, resulting in a combined volume of 1.55 mL of nutrient solution and 6.0M saltwater being added every hour. At hour ~248, on experimental-day 10.5, the target molarity is now 1.4M. Since nutrient solution is still being added at a rate of 1.5 ml/hr, the script will add 0.45 ml of 6.0M saltwater every hour. The program will calculate the amount of saltwater to be added every hour. There is a little bit of logic to calculate the saltwater dose volume and dosing intervals to avoid impracticalities, but the important part takeaway is the rate of saltwater being added per hour.
Temperature Schedule: The temperature changes every 8 hours following this schedule: [21C → 28C → 34C → 37C → 34C → 28C → repeat from beginning]
I also manually started the stirrer at 600 rpm and the od600/growth/etc automations at the beginning of the experiment.
Experimental Setup
I used 3 glass jars (250ml) to hold the media solution, saltwater solution, and waste. First, I sterilized all glass jars, as well as the chamber/cap/stir bar (unscrewed, but cap resting on top), by boiling water in a large cooking pot with a glass lid on top. Once I was satisfied, I turned off the heat and kept the glass lid on, and removed the glassware as needed.
For the saltwater solution, I weighed out 87.44g of salt and 252g of tap water, and mixed them. Initially, I was targeting a 6.0M solution (just below the saturation point of ~6.1M at 20C). My roommates helped a bit, and while trying to get the salt to dissolve, I’m pretty sure water and/or salt was added/evaporated.
In the future, I think a better approach would be:
- Decide on using supersaturated saltwater (6.1M) instead of almost supersaturated saltwater (6.0M)
- Calculate the desired volume of saltwater, and then weigh out that much water.
- Calculate and weigh out enough salt to supersaturate the amount of water. Add in a little extra to ensure saturation.
- Pour the water into a coffee mug, and heat the water to a boil in the microwave (watch the microwave the entire time).
- Mix the salt into the heated water until fully dissolved. No solid salt should remain.
- You should have an excess of saltwater by volume, so weigh out the desired volume of saltwater. (keep in mind, saturated saltwater has a density of 1.2g/ml)
- Pour the saturated saltwater into the glass jar. Done.
Nutrient Solution: The beef broth recommended mixing each packet (15g) with 10 oz of hot water. I sterilized the water by boiling ~270g water (can use microwave) and mixing in the beef broth while still hot. With the beef broth still near-boiling, I added it to the nutrient jar, hoping that by adding the broth to the jar while still near-boiling, it would be self-sterilizing until I capped the jar.
A picture of my setup.
I mixed the yeast powder in room-temperature water I had previously boiled and then used a small syringe (sterilized with 70%ISO) to transfer ~ 3ml of yeast-water to the pio-chamber. I then filled the pio-chamber to running volume with nutrient media. Then, I started the experiment.
Ideas for improvement
- Find a better way to prepare and store nutrient/salt solutions. It is difficult and a hassle to sterilize it and my current setup doesn’t allow me to easily sterilize or store it. I plan on using IV bags or plant watering bags to store my solutions. I could prepare the solutions ahead of time, sterilize them by placing them in water heated to a boil, store them in the freezer, and then unthaw when I run the next experiment.
- I’ll add in more later as I think of them
Test Code
from pioreactor.background_jobs.dosing_control import DosingController
from pioreactor.background_jobs.temperature_control import TemperatureController
from pioreactor.utils.timing import RepeatedTimer
from pioreactor.whoami import get_unit_name
class Schedule():
def __init__(self, salt_value_array, dose_value_array, temp_value_array, **kwargs):
self.salt_value_array = salt_value_array
self.dose_value_array = dose_value_array
self.temp_value_array = temp_value_array
self.dose_state_array = {
'run_flag': True, 'run_interval': 20, 'run_counter': 0,
'update_flag': True, 'update_interval': 60, 'update_counter': 0,
'run_target': 0.5, 'previous_run_amount': 0, 'name': 'nutrient_media',
'interval_counter': 0}
self.salt_state_array = {
'run_flag': True, 'run_interval': 20, 'run_counter': 0,
'update_flag': True, 'update_interval': 720, 'update_counter': 0,
'run_target': 0.1, 'previous_run_amount': 0, 'name': 'salt_media',
'interval_counter': 0} # 720 update interval -> 12 hours
self.temp_state_array = {
'run_flag': True, 'run_interval': 20, 'run_counter': 0,
'update_flag': True, 'update_interval': 480, 'update_counter': 0,
'run_target': 28, 'previous_run_amount': 0, 'name': 'temp_control',
'interval_counter': 0} # 480 update interval -> 8 hours
def check_for_updates(self, state_array):
if counter - state_array['run_counter'] >= state_array['run_interval']:
state_array['run_flag'] = True
state_array['run_counter'] = counter
if counter - state_array['update_counter'] >= state_array['update_interval']:
state_array['update_flag'] = True
state_array['update_counter'] = counter
return state_array
def update_target_values(self, state_array):
if state_array['update_flag'] == True:
if state_array['name'] == 'salt_media':
target_molarity = self.salt_value_array[state_array['interval_counter']]
if target_molarity == 0:
state_array['run_target'] = 0
elif target_molarity > 0:
values = self.calc_salt_dosing(target_molarity)
state_array['run_target'] = values[0]
state_array['run_interval'] = values[1]
state_array['interval_counter'] += 1
state_array['interval_counter'] %= (len(self.salt_value_array)+1) # loop around to begining to prevent out-of-bounds
elif state_array['name'] == 'nutrient_media':
state_array['run_target'] = state_array['run_target'] # Not currently in use.
elif state_array['name'] == 'temp_control':
state_array['run_target'] = self.temp_value_array[state_array['interval_counter']]
state_array['interval_counter'] += 1
state_array['interval_counter'] %= (len(self.temp_value_array)+1)
state_array['update_flag'] = False
return state_array
def get_run_values(self):
media_ml = 0
alt_media_ml = 0
waste_ml = 0
temp_target = 0
if self.dose_state_array['run_flag'] == True:
media_ml = self.dose_state_array['run_target']
self.dose_state_array['run_flag'] = False
if self.salt_state_array['run_flag'] == True:
alt_media_ml = self.salt_state_array['run_target']
self.salt_state_array['run_flag'] = False
if self.temp_state_array['run_flag'] == True:
temp_target = self.temp_state_array['run_target']
temp_target = self.temp_state_array['run_target']
waste_ml = media_ml + alt_media_ml
run_value_settings = {'media_ml': media_ml, 'alt_media_ml': alt_media_ml, 'waste_ml': waste_ml, 'temp_target': temp_target}
return run_value_settings
def calc_salt_dosing(self, diluted_molarity):
# takes a target molarity and returns saltwater dose volume and inter [minutes/dose]
# Settings assume 6.0 molarity NaCl-water solution
solution_molarity = 6.0 # NaCl saturation point ~= 6.1 molarity in 20C H2O
salt_dose_volume = 0.1 # Starting dose volume
fraction = diluted_molarity/(solution_molarity - diluted_molarity) # V_salt = V_Nut * M_dil / (M_NaCl - M_dil) = V_Nut * fraction
nutrient_volume_per_hour = 60*self.dose_state_array['run_target']/self.dose_state_array['run_interval']
salt_volume_per_hour = nutrient_volume_per_hour * fraction
salt_dose_interval = round(60*salt_dose_volume/salt_volume_per_hour)
while salt_dose_interval < 20:
salt_dose_volume += 0.1
salt_dose_interval = round(60*salt_dose_volume/salt_volume_per_hour)
values = [salt_dose_volume, salt_dose_interval]
return values
def run_execute(self):
self.dose_state_array = self.check_for_updates(self.dose_state_array)
self.dose_state_array = self.update_target_values(self.dose_state_array)
self.salt_state_array = self.check_for_updates(self.salt_state_array)
self.salt_state_array = self.update_target_values(self.salt_state_array)
self.temp_state_array = self.check_for_updates(self.temp_state_array)
self.temp_state_array = self.update_target_values(self.temp_state_array)
run_value_settings = self.get_run_values()
return run_value_settings
# Lists of setpoints for dosing, salting, and temperaturing
dose_value_array=[0.5]
salt_value_array=[0, 0.2, 0, 0.4, 0, 0.6, 0, 0.8, 0, 1, 0, 1.2, 0, 1.4, 0, 1.6, 0, 1.8, 0, 2, 0, 2.2, 0, 2.4, 0, 2.6, 0, 2.8, 0, 3, 0, 3.2, 0, 3.4, 0, 3.6]
temp_value_array=[21, 28, 34, 37, 34, 28]
sc = Schedule(salt_value_array, dose_value_array, temp_value_array)
# "silent" automation is basically: do nothing. We'll manually invoke dosing later.
dc = DosingController(
"silent",
duration=None,
unit=get_unit_name(),
experiment="salt_temp_cycle_experiment"
)
tc = TemperatureController(
"thermostat",
target_temperature=30,
unit=get_unit_name(),
experiment="salt_temp_cycle_experiment"
)
SCHEDULE = {
1: {'media': 0.5, 'alt_media': 0.0, 'temperature': 35},
2: {'media': 0.5, 'alt_media': 0.0, 'temperature': 35}
}
counter = 0
previous_temp = 0
def update():
global counter
global previous_temp
run_value_settings = sc.run_execute()
temperature = run_value_settings['temp_target']
media_per_dose = run_value_settings['media_ml']
alt_media_per_dose = run_value_settings['alt_media_ml']
waste_ml = run_value_settings['waste_ml']
if temperature is not previous_temp:
tc.automation_job.set_target_temperature(temperature)
previous_temp = temperature
dc.automation_job.execute_io_action(alt_media_ml=alt_media_per_dose, media_ml=media_per_dose, waste_ml = waste_ml)
counter += 1
# this RepeatedTimer will run `update` every 60 seconds
scheduler = RepeatedTimer(60, update).start()
# block here
dc.block_until_disconnected()