Hello everyone.
I am having problems using the pioreactor for experiments after doing the calibration for the OD600 measurements. So the calibration went fine ( i used puTTy to access the Rasberry pi and everything worked out well with a nice calibration curve in the end from OD600 = 0.1 to OD600 = 1.0) but when i start my new experiment and I want to start the OD measurements it doesn’t work, it doesn’t even give me an error response. It just tries to connect and fails so i cannot run my experiments.
Do you have any idea how to solve this problem?
this is what is says in the config.ini file:
#use the most recent calibration file, if available
use_calibration=1
I called my calibration 1.
If i switch it back to use_calibration=0 it works out but i would like to use my calibration file.
Greetings
Leo
Hi @LeoKleym! Welcome to the forum!
Let’s see. Does OD Reading work if use_calibration=0? Or does OD Reading just never start, regardless?
One thing to try: use PuTTY to access the specific Raspberry Pi again, and run the following:
pio run od_reading
This should provide more information about if it fails, and how it fails.
Hey @CamDavidsonPilon !
Thanks for the fast reply. It does work if i am not using my calibration file, so if i use calibration=0.
Any ideas why it won’t use my calibration file?
It could be a schema-mismatch. We did change the schema in the last update (but this should have been automatically fixed for you). Can you try the suggestion of setting use_calibration=1 in the config and running pio run od_reading on the command line?
i changed the line in the confic to use_calibration=1 and then i run the
pio run od_reading the following comes up:
pioreactor@pioreactor1:~ $ sudo pio run od_reading
2023-07-12T08:49:51+0100 DEBUG [ir_led_ref] Using PD channel 1 as IR LED reference.
2023-07-12T08:49:52+0100 DEBUG [calibration_transformer] Using calibration calibration1 for channel 2
2023-07-12T08:49:52+0100 DEBUG [od_reading] Init.
2023-07-12T08:49:52+0100 DEBUG [od_reading] Starting od_reading with PD channels {‘2’: ‘90’}, with IR LED intensity 50.0% from channel A.
2023-07-12T08:49:53+0100 DEBUG [adc_reader] Using ADC class Pico_ADC.
2023-07-12T08:49:53+0100 DEBUG [adc_reader] ADC ready to read from PD channels 2, 1, with gain 1.
2023-07-12T08:49:54+0100 DEBUG [adc_reader] AC hz estimate: 50.0
2023-07-12T08:49:54+0100 DEBUG [adc_reader] ADC offsets: {‘2’: 1252, ‘1’: 1247}, and in voltage: {‘2’: 0.06305860805860806, ‘1’: 0.06280677655677655}
2023-07-12T08:49:54+0100 INFO [od_reading] Ready.
2023-07-12T08:49:54+0100 DEBUG [od_reading] od_reading is blocking until disconnected.
So here it seems like its working! I will run another experiment tomorrow and check if it works…But i can still not turn it on over the surface UI, any ideas why it works via puTTY but not over the UI? Everything else is working out fine.
thanks a lot for your help!
Oh, that’s unexpected. Do other jobs start correctly from the UI, like stirring for example? (I guess the answer is yes)
Let me investigate this some more. For now, to make sure the job doesn’t disconnect if putty disconnects, you can use something like:
pio run od_reading >/dev/null 2>&1 & disown
One thing that would help me to debug is the following. Have use_calbration=1, and attempt to start the OD job from the UI. Probably it will not start.
Next, use Putty to access the Raspberry Pi, and type:
pio logs
This is a stream of recent logs. I am wondering if you see a line like:
[monitor] DEBUG Running `nohup pio run od_reading >/dev/null 2>&1 &` from Monitor job.
Using this command
pio run od_reading >/dev/null 2>&1 & disown
helped for the OD reading not to disconnect when puTTY disconnects.
If i want to start any other job from the UI it works, just the OD reading won’t start. Stopping it over the UI works though…
in the pio logs follwing lina came up:
2023-07-13T10:40:54+0100 [monitor] DEBUG Running nohup pio run od_reading >/dev /null 2>&1 & from Monitor job.
Weird! I’m a bit stumped.
Here’s how it should work:
- You click the Start button in the UI. This sends a request to the web UI’s backend
- The backend sends a message to the Pioreactor’s
monitor job. This job will perform essentially a “headless” version of you writing pio run od_reading >/dev/null 2>&1 & disown on the command line. This is the log output you observed.
- The job starts, and the UI flips to “Stop” button.
So for some reason, the job is failing to start at step 3.
Can you post your Pioreactor’s version output?
pio version -v
Sure!
here it is:
Pioreactor software: 23.6.27
Pioreactor HAT: 1.2
Pioreactor firmware: 0.2
HAT serial number: ec265e18-ea3c-4a58-a3a9-01b6c024c8bb
Operating system: Linux-6.1.21-v7±armv7l-with-glibc2.31
Raspberry Pi: Raspberry Pi 3 Model A Plus Rev 1.0
Image version: ad3ac2e012668dba2280c00cdf288b7ac66cbfca
Pioreactor UI: 23.6.26
1 Like
So I don’t really have a solution yet, but I added some error handling around this workflow to the develop branch. More error logs (if any) should appear in pio logs when you try to start the job from the UI.
To upgrade to try this branch, on the Pioreactor:
pio update app -b develop
And then, if you want to switch back to the latest stable version:
pio update app -v 23.6.27
Hello again,
so I am still having the same issues with the ODcalibration files but with the
pio run od_reading >/dev/null 2>&1 & disown
it’s working fine.
I just got another question, so I calibrated one of my Pios and i got a cluster now consisting of 5 Pioreactors with one leader which is also working as a worker. Is it possible to just calibrate one Pioreactor and to use this file for the whole cluster or do i need to calibrate each pioreactor seperately?
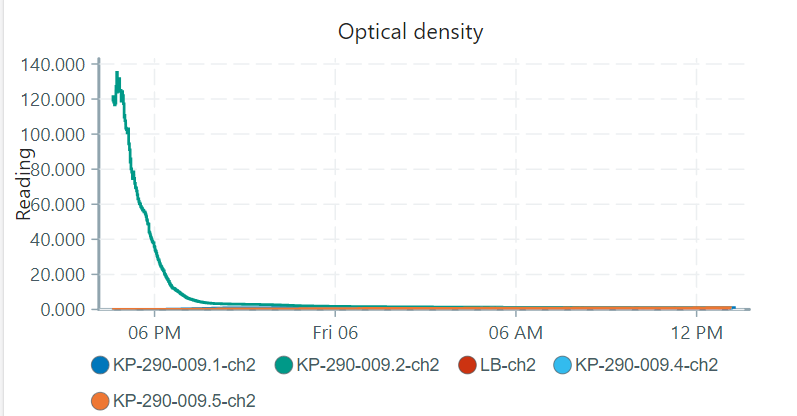
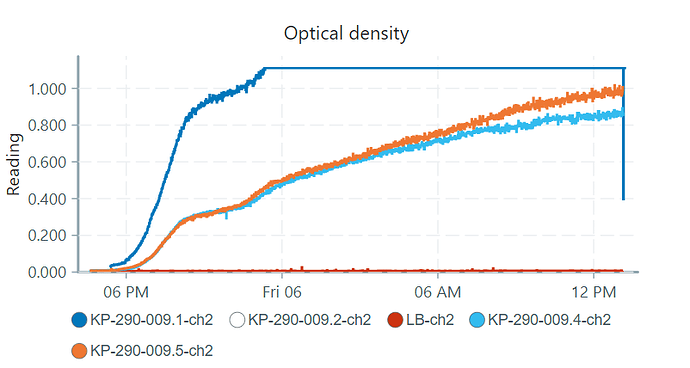
Also, another weird thing came up. one of my pioreactors is reading the OD’s definitely wrong because i always get a negative output:
compared to the other pio’s which had excacly the same bacterial solution with the same starting OD in them:
(Just as a remark: KP-290-009.1 (pioreactor 1) seems to have different values because this is the one i calibrated, the others i did not calibrate yet.)
Greetings and it’s fun working with the cluster!
Leo
Hi Leo,
Due to the variations between the Pioreactors (ex: slightly different IR LEDs and PDs via manufacturing variations), I recommend calibrating each one independently. Sharing calibrations is something we just never tried. However, calibrations are just files on disk, so you can share them between units if you really want.
This looks like backwards PDs. On that unit (KP290-009.2), can you confirm that PD2 on the HAT is connected to the 90deg pocket?
Hey! Hah yes, the cables where switched on my one Pioreactor…
So i am wondering if user users are also experiencing this:
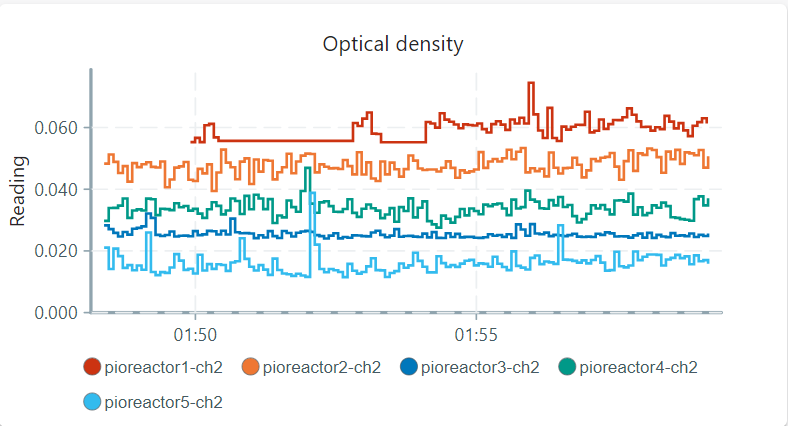
I calibrated all pioreactors with the same bacterial strain and Media and OD’s, from 0.03 - 1.5 OD600. What I am seeing now if i compare the reading of the pioreactors with each other is that there are differences in OD measurements even though i prepared 100 ml of my LB + bacteria at OD600 = 0.04 and alliquotted it into the vials, 14 ml each. So it is excacly the same in each vials but the OD readings look like this:
you got any ideas on how to make them run the same? I want to perform experiments where it is very important that the OD measurements are accurate, I would be happy if you could give me some advice on how to do that or even if it is possible with the pioreactor. I calibrated them with an OD600 starting at 1.5 and always added 1 ml of my Media, I said that my Media’s OD is 0.0 because i already blanked my Spectrophotometer with it and I always use LB anyways.
Greetings!
Leo
It’s always going to be hard to get perfect concordance between the Pioreactors. For example, even differences in vial glass thickness will be a uncontrollable parameter. However, the amount of variance you are seeing is beyond what we like.
One thing you can try to help narrow down where the major source of variance is located: try preparing a simple solution in a single vial, and pass that between the Pioreactors (with stirring and OD on). Keep the vial in the same orientation relative to the IR LED (i.e. don’t allow the vial to rotate). In theory, the ODs should be nearly the same, but I’m curious how much variance you observe.